Every cancer is unique, but many share common genetic drivers and molecular triggers that fuel tumor growth, regardless of their tissue of origin. Pan-cancer analysis uncovers genetic alterations that cut across cancer types, revealing opportunities to use the same therapy for tumors in different organs when they share the same DNA driver. This shifts the focus from tumor location to root genetic drivers paving the way for truly personalized, biology-driven treatment. A pan-cancer panel powered by next-generation sequencing (NGS) is designed to look beyond the boundaries of a single tumor type. Instead, it analyzes a broad set of clinically relevant genes to capture the full spectrum of cancer-associated alterations. This tumor-agnostic approach identifies multiple classes of genomic changes from single nucleotide variants (SNVs) and insertions/deletions (indels) to copy number variations (CNVs), microsatellite instability (MSI), and tumor mutational burden (TMB).
By detecting these critical drivers across diverse cancers, pan-cancer panels provide insights that can guide precision treatment decisions, match patients with targeted therapies or immunotherapies, and support clinical trial enrollment. This comprehensive view makes pan-cancer testing a powerful tool in advancing personalized oncology care.
Genes2Me PanCan is a CE-IVD certified in vitro diagnostic comprehensive genomic profiling NGS assay aimed to screen a range of cancer-causing genes to identify somatic mutations in DNA & RNA from human clinical samples like FFPE and fresh tissue covering all the coding sequences enriched by hybridization capture-based target enrichment. It captures all major variant types along with key immuno-oncology biomarkers such as Microsatellite Instability (MSI) and Tumor Mutational Burden (TMB). In addition, the panel is equipped to detect Epstein-Barr Virus (EBV) and Human Papilloma Virus (HPV), offering valuable insights into virus-associated cancers. With coverage of approximately 681 genes, including coding sequences (CDS), hotspot regions, and fusion genes, the panel spans a target size of nearly 1.7 Mb, ensuring broad and clinically relevant genomic profiling. The panel uncovers critical mutations driving tumor progression and therapy resistance, opening new avenues for precision-guided therapies in solid cancers.
| Cancer types | Genes |
|---|---|
| Lung Cancer | EGFR, KRAS, ALK, RET, PIK3CA, EGFR, KRAS, MET, PTEN, RET, BRAF, ERBB2, ALK |
| Breast Cancer | BRCA 1, BRCA 2, PTEN, TP53, CHEK2, BRIP1, ATM, PALB2, PIK3CA, PMS2, ESR1, FGFR1 |
| Colorectal Cancer | RAS, EGFR, TGF, ATM, BRAF, CHEK2, NRAS, PIK3CA, PTEN, TP53 |
| Ovarian Cancer | CHEK2, TP53, BARD1, KRAS, RAD51, BRIP1, PALB2, BRAF, ERBB2, PTEN, PIK3CA, BRCA1 |
| Oesophagus Cancer | ERBB2, EGFR, RB1 |
| Bladder Cancer | TP53, RB1, HRAS, PIK3CA, FGFR3, ATM, MTOR |
| Prostate Cancer | BRCA 1, HOXB13, AR, ATM, MYC, PTEN, RAF1, BRCA2 |
| Pancreatic Cancer | BRCA 1, BRCA 2, EGFR, HRAS, KRAS, PALB2, PIK3CA, TP53 |
| Liver Cancer | TP53, CDKN2A |
| Gastric Cancer | APC, MLH1, MSH2, MSH6, EPCAM |
| Thyroid Cancer | BRAF, RAS, RET, TP53, PTEN |
| Cervical Cancer | DICER1, MED1, HLA-A, PI3K, MAPK |
Coverage uniformity for the G2M Pan-Cancer CGP Panel
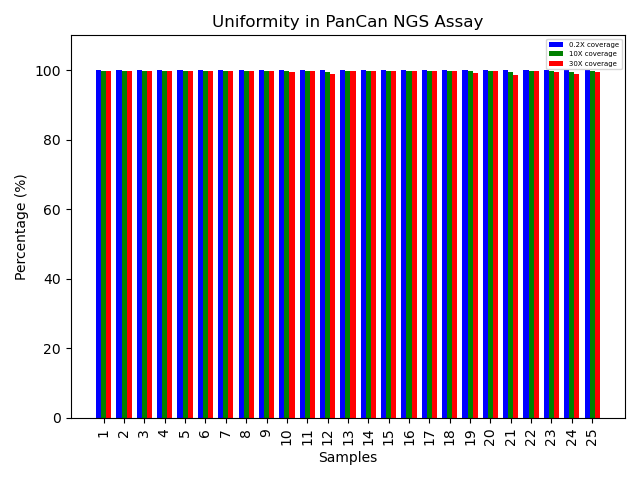
Greater than 0.2X mean coverage is observed for >99% of targets. The genomic DNA libraries from blood and FFPE tissues (n=64) were enriched using the G2M Pan-Cancer NGS Panel and sequenced on a NovaSeq system using 2 x 150 paired-end reads. The data represents near-complete uniform coverage (~100%) across all samples at 0.2X, 10X, and 30X thresholds. This high level of uniformity indicates that the probe design is well optimized, enabling targeted NGS assays to consistently deliver high-confidence and reproducible results.
From FFPE to Blood - G2M PanCancer CGP Panel Ensures Consistent Performance Where It Matters Most.
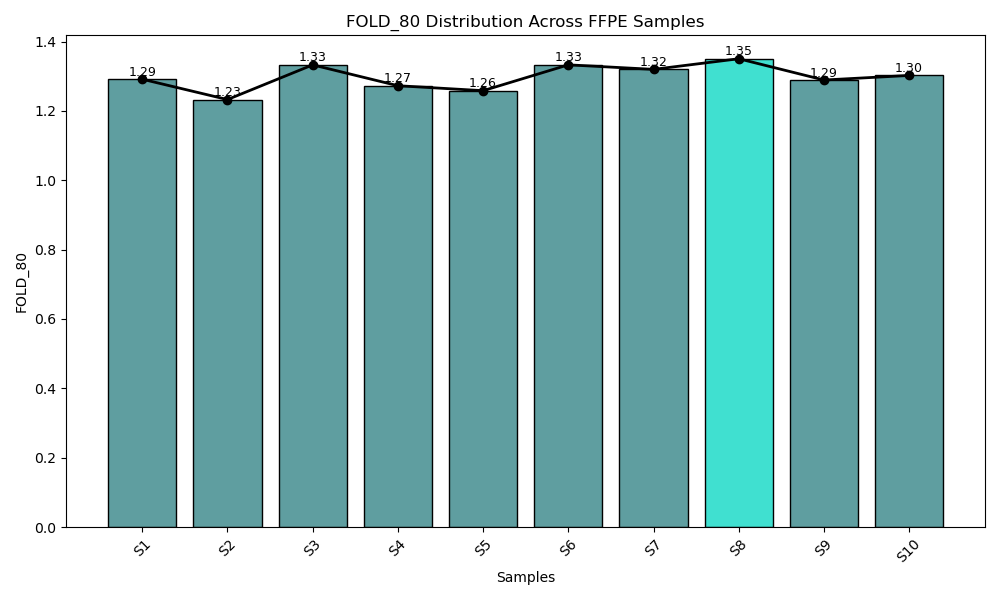
Consistent fold 80 base penalty across both FFPE (blue bar) and blood-derived (cyan bar) samples. The G2M Pan-Can CGP Panel demonstrates a fold 80 base penalty below 1.3, reflecting exceptional coverage uniformity across target regions. This low value indicates that only 1.3× the average sequencing depth is required to achieve 80% coverage at the desired threshold, minimizing over-sequencing and ensuring efficient, balanced read distribution affirming the robustness of the data quality.
Mean & Median Gene Coverage Supporting Clinical Reliability
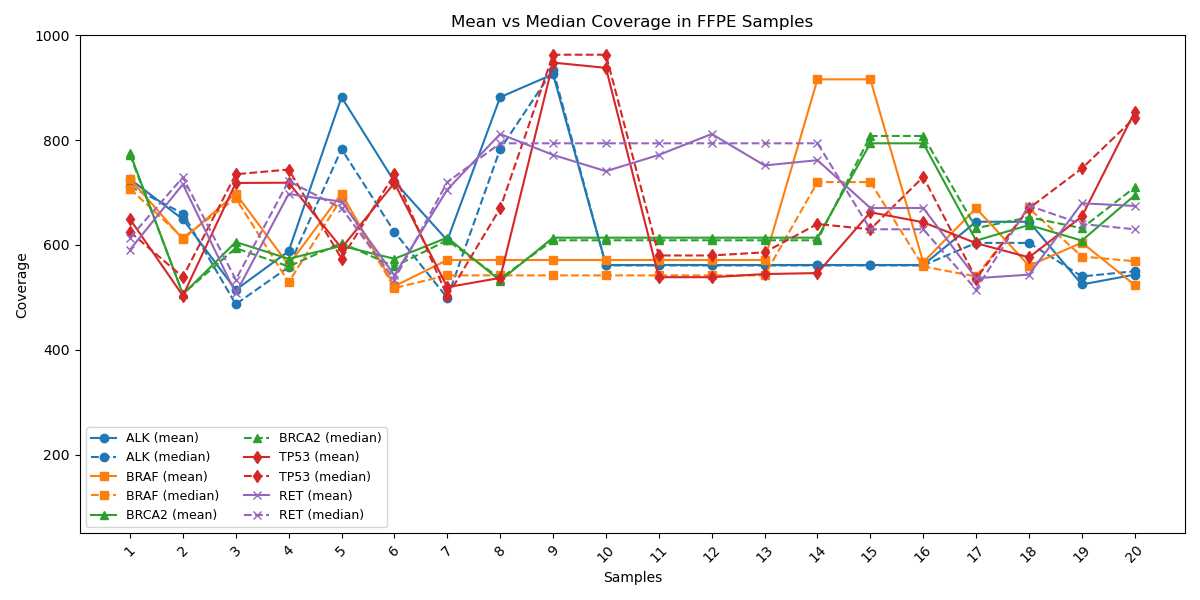
Coverage profiles of key cancer-associated genes (ALK, BRAF, BRCA2, TP53, RET) are shown with mean (solid line) and median (dashed line) values. Displaying both mean and median coverage metrics provides a more complete picture of sequencing performance, as each highlights different aspects of data uniformity and reliability. A close alignment between mean and median coverage in NGS assays.
On-Target Alignment in Cancer Samples
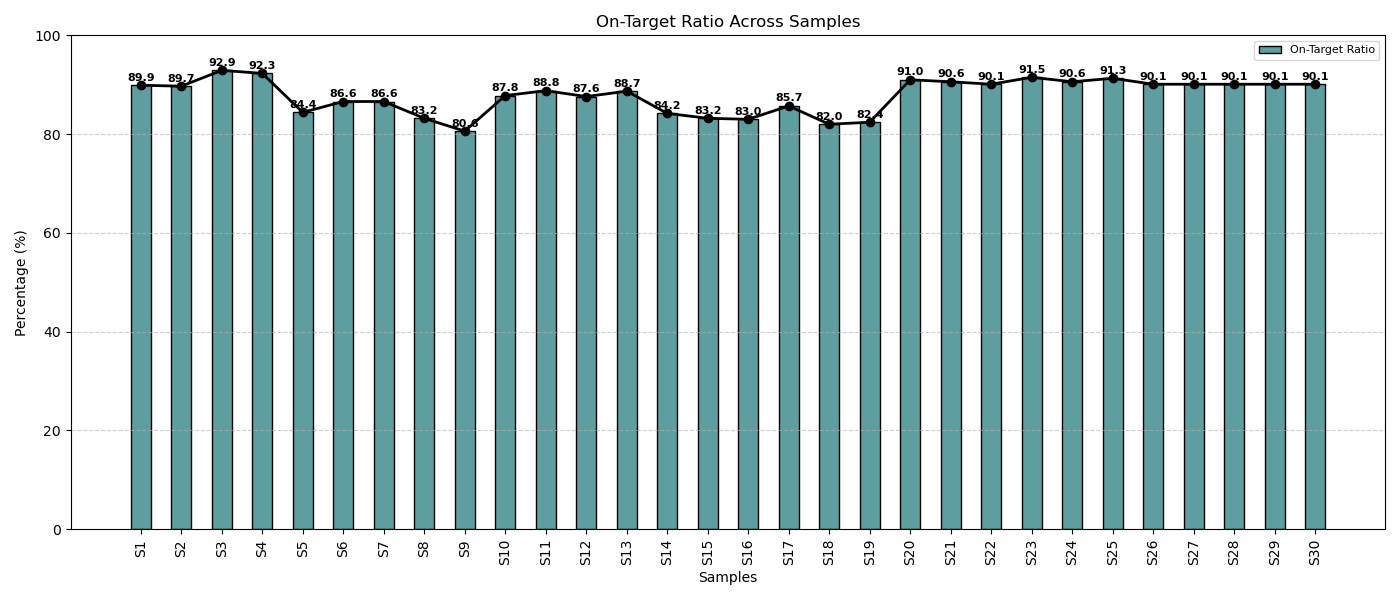
Across all cancer patient samples, the on-target alignment consistently exceeded 80%, reflecting the panel’s high specificity, optimized assay conditions, and efficient target enrichment. This strong performance underscores the reliability and precision of the sequencing workflow.
| Commercial Name | Old Cat No. | New Cat No. | Pack Size | Platform |
|---|---|---|---|---|
| PanCan CGP NGS Test Kit | G2MCP06001 | G710006-1 | 24 T | Illumina |
| G2MCP06001 | G710006-2 | 96 T | Illumina | |
| G2MCP06001 | G710006-3 | 96 T - EZY | Illumina – EZY | |
| PanCan CGP NGS Test Kit | G2MCP06001 | G710006-4 | 24 T | MGI |
| G2MCP06001 | G710006-5 | 96 T | MGI | |
| G2MCP06001 | G710006-6 | 96 T – EZY | MGI – EZY | |
| PanCan CGP NGS Test Kit | G2MCP06001 | G710006-7 | 24 T | Aviti |
| G2MCP06001 | G710006-8 | 96 T | Aviti | |
| G2MCP06001 | G710006-9 | 96 T – EZY | Aviti – EZY | |
| PanCan CGP NGS Test Kit | G2MCP06001 | G710006-10 | 24 T | Thermo |
| G2MCP06001 | G710006-11 | 96 T | Thermo | |
| G2MCP06001 | G710006-12 | 96 T – EZY | Thermo – EZY |
Since its inception in 2016, Genes2me has been constantly striving towards setting a benchmark in the diagnostics space by introducing premium quality (Made in India) diagnostic kits which are CE-IVD, ISO-13485:2016, and ISO 9001:2015 certified, assuring our clients of unparalleled quality and compliance with international standards.
© 2025 Genes2me. All rights reserved.