The Lymphoma NGS Assay is a hybridization capture–based next-generation sequencing assay designed for comprehensive molecular profiling of lymphoid malignancies. The assay enables high-resolution detection of clinically relevant genomic alterations to support lymphoma subtype classification, prognostic stratification, and identification of actionable biomarkers for targeted therapy selection.
The Lymphoma NGS Assay combines DNA- and RNA-based analyses within a single workflow, supporting histopathological evaluation and facilitating precision diagnostics consistent with WHO and ICC lymphoma classification guidelines.
Lymphomas are driven by a heterogeneous spectrum of genomic alterations arising from aberrant somatic hypermutation, dysregulated antigen receptor signaling, epigenetic deregulation, and recurrent chromosomal rearrangements. Key oncogenic mechanisms include:
Accurate detection of these molecular events is essential for lymphoma subclassification, prognostic assessment, and therapeutic decision-making. The Lymphoma NGS Assay is engineered to deliver uniform and sensitive coverage across biologically complex regions commonly implicated in lymphoid malignancies.
The Lymphoma NGS Assay is designed for comprehensive genomic coverage, encompassing 134 genes associated with DNA mutations, 21 DNA fusion genes, and 94 RNA fusion genes within a total target size of approximately 0.7 Mb. The assay covers entire coding sequences along with clinically relevant hotspot regions, enabling robust detection of disease-defining and actionable variants. This integrated assay architecture supports simultaneous interrogation of single nucleotide variants, small insertions and deletions, copy number variations, and both DNA- and RNA-based fusion events within a single, consolidated assay workflow.
The assay employs a hybridization capture–based target enrichment strategy, engineered for efficient and uniform capture of complex genomic regions, including GC-rich, repetitive, and homologous sequences. An optimized hybridization time of approximately 4 hours enables rapid library preparation without compromising to capture specificity or depth of coverage.
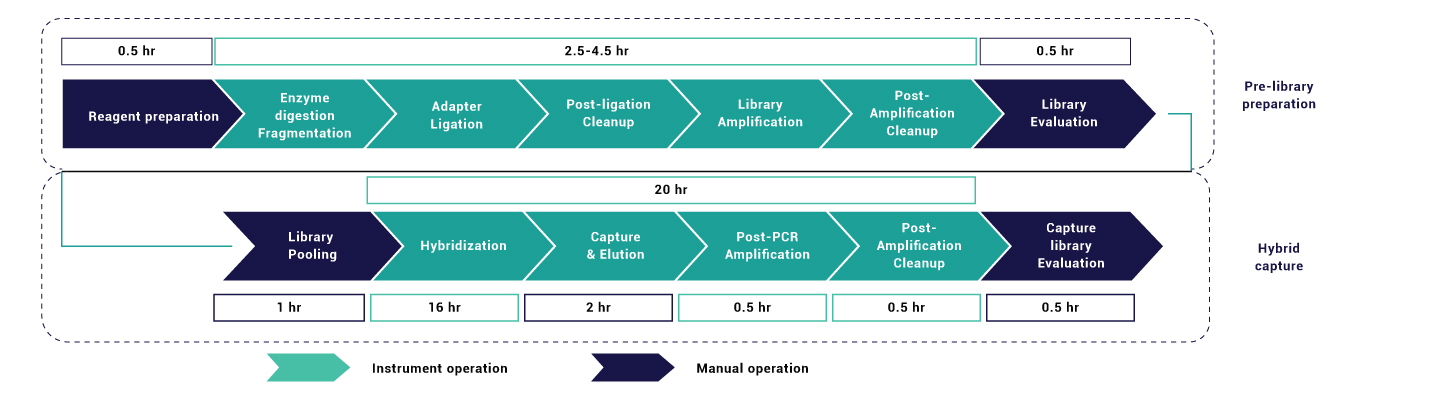
The workflow is designed to be automation-friendly, supporting both manual and automated library preparation formats to accommodate varying laboratory throughput requirements. The assay demonstrates platform-agnostic compatibility and has been validated across commonly used sequencing systems, including Illumina, Element Biosciences, MGI, and Thermo Fisher platforms. This streamlined and flexible workflow ensures high reproducibility, consistent assay performance, and scalability across diverse clinical laboratory settings.
NGS data analysis is supported by GATK-based pipelines integrated with the Cliseq Interpreter Platform, a cloud-based clinical interpretation solution designed to streamline analysis of complex genomic data.
This integrated bioinformatics framework enables standardized, reproducible, and clinically actionable interpretation of lymphoma genomic data.
Exceptional on-target performance across lymphoma samples
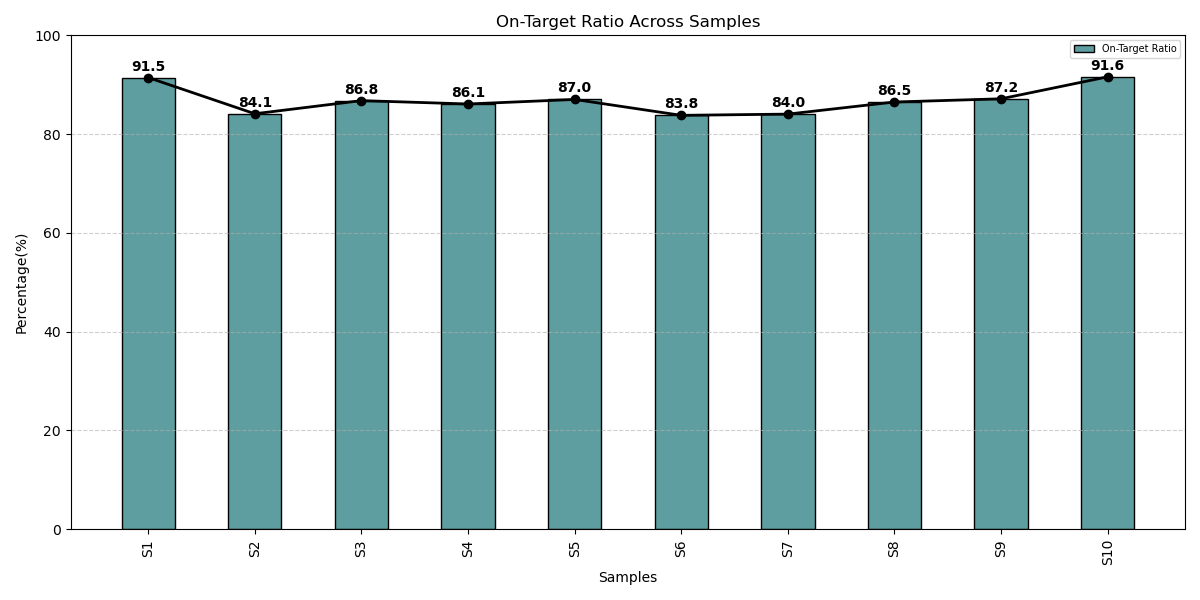
All patient samples demonstrated over 80% on-target alignment reflecting the Assay’s precision engineered probe architecture and rigorously optimized assay chemistry. This high capture efficiency ensures uniform coverage across target regions, enabling reproducible, high confidence variant detection essential for clinical grade sequencing and scalable diagnostic workflows.
Balanced coverage of critical genes reflects robust workflow
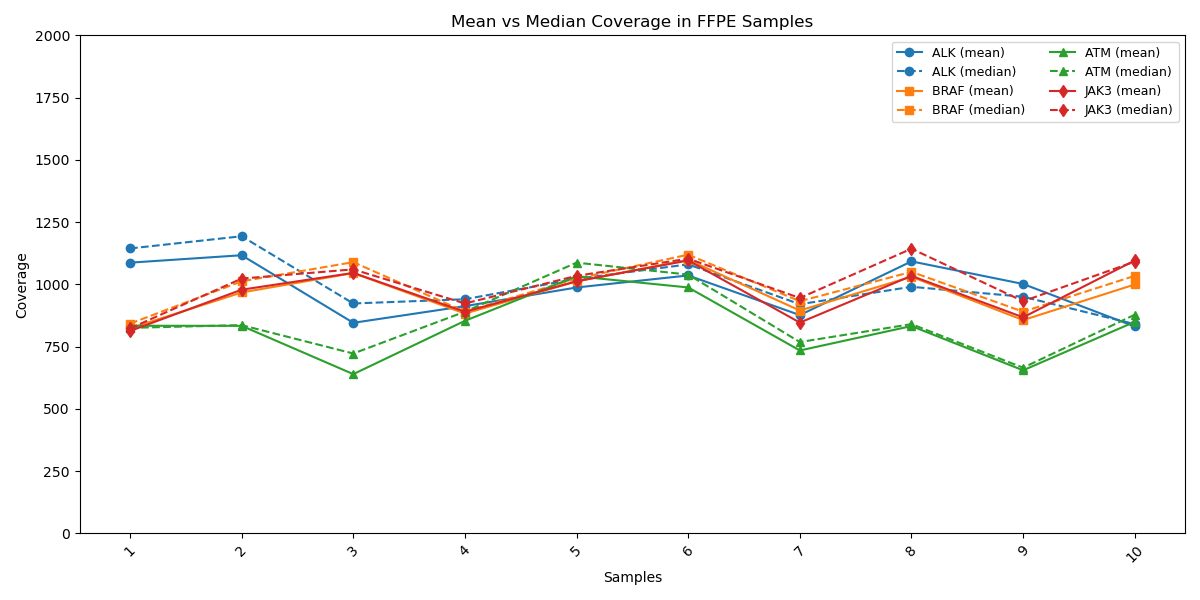
Coverage profiles for critical genes such as ALK, BRAF, ATM, and JAK3 showing near perfect alignment between mean (solid line) and median (dashed line) depth. This tight concordance reflects the assay’s uniform performance across all target regions, minimizing bias and ensuring robust, reproducible results. Such consistency is vital for confidently interpreting genomic data across diverse FFPE samples, reinforcing the assay’s value in precision oncology.
ALK centered fusion map highlighting major partner genes
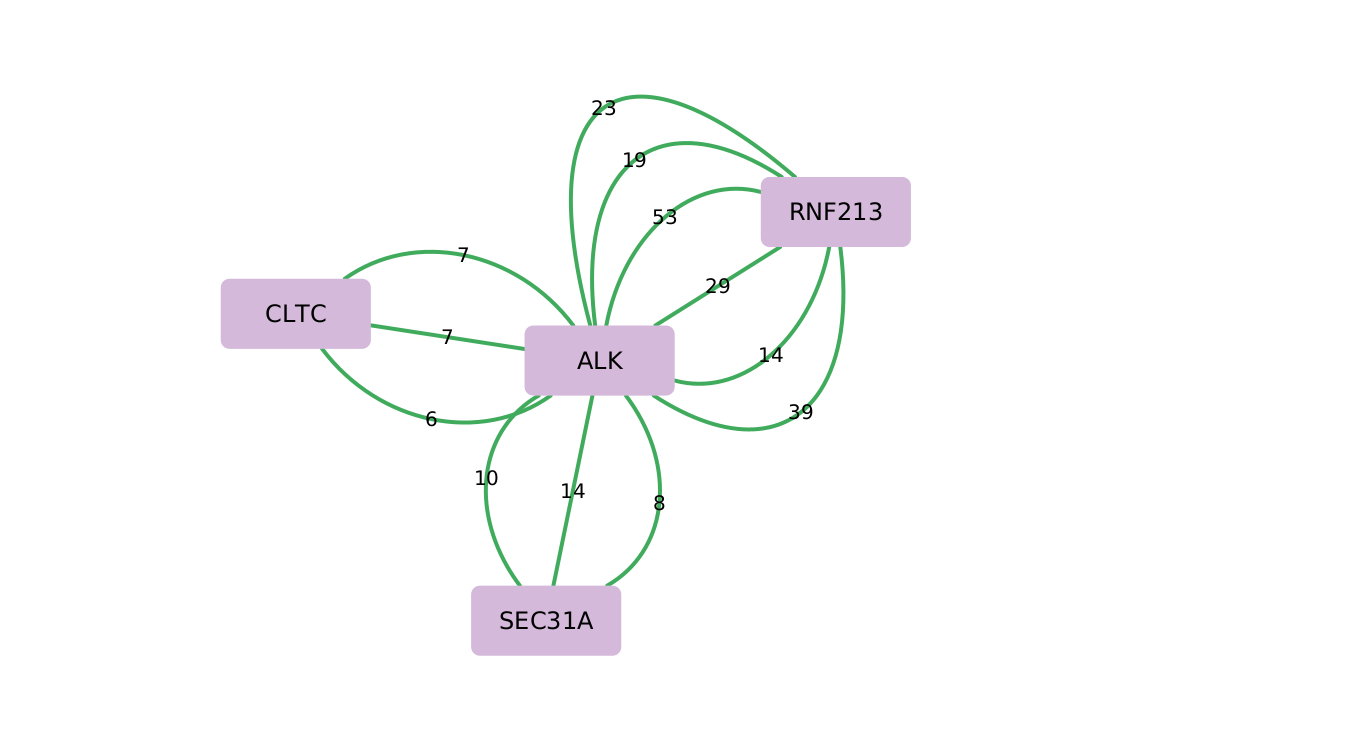
DNA fusions identified with G2M Lymphoma NGS Assay in plasma samples along with support reads.
Mutation landscape of top lymphoma genes
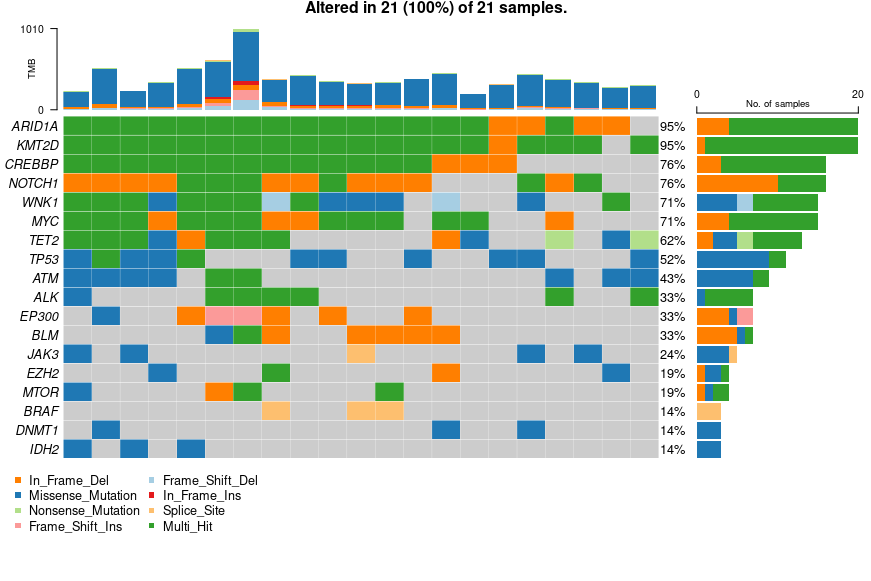
The Oncoplot shows the somatic mutation landscape of the top 18 lymphoma associated genes. Missense variants (blue) dominate, while multi-hit events (green) indicate multiple mutation types within the same gene. The right panel shows mutation frequency per gene.
| Commercial Name | Old Cat No. | New Cat No. | Pack Size | Platform |
|---|---|---|---|---|
| Lymphoma NGS Test Kit | G2MLYM31001 | G710018-1 | 24 T | Illumina |
| G2MLYM31001 | G710018-2 | 96 T | Illumina | |
| G2MLYM31001 | G710018-3 | 96 T – EZY | Illumina – EZY | |
| Lymphoma NGS Test Kit | G2MLYM31001 | G710018-4 | 24 T | MGI |
| G2MLYM31001 | G710018-5 | 96 T | MGI | |
| G2MLYM31001 | G710018-6 | 96 T – EZY | MGI – EZY | |
| Lymphoma NGS Test Kit | G2MLYM31001 | G710018-7 | 24 T | Aviti |
| G2MLYM31001 | G710018-8 | 96 T | Aviti | |
| G2MLYM31001 | G710018-9 | 96 T – EZY | Aviti – EZY | |
| Lymphoma NGS Test Kit | G2MLYM31001 | G710018-10 | 24 T | Thermo |
| G2MLYM31001 | G710018-11 | 96 T | Thermo | |
| G2MLYM31001 | G710018-12 | 96 T – EZY | Thermo – EZY |
Since its inception in 2016, Genes2me has been constantly striving towards setting a benchmark in the diagnostics space by introducing premium quality (Made in India) diagnostic kits which are CE-IVD, ISO-13485:2016, and ISO 9001:2015 certified, assuring our clients of unparalleled quality and compliance with international standards.
© 2025 Genes2me. All rights reserved.