High Confidence Gene Annotation Across Trusted Databases
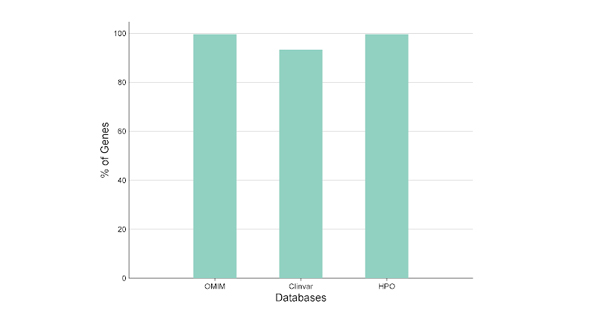
Reference coverage from curated sources (percentage of genes from each database).
The human genome consists of DNA sequences organized into coding regions, known as exons, and non-coding segments, called introns. Exons collectively make up the exome, which encodes proteins essential for biological functions. Clinical exome sequencing (CES) involves analyzing these coding regions to identify pathogenic variants, especially in cases where the condition is complex or present in an unusual way. This approach holds significant medical value, as it enables the detection of disease-causing mutations across a wide spectrum of genetic disorders. The test focuses on genes scientifically linked to diseases, carefully curated from trusted databases including OMIM, HGMD, and ClinVar.
By identifying disease-causing variants, it enables clinicians to determine the most suitable treatment strategies at the earliest stage, helping to manage symptoms effectively and improve overall quality of life. It is particularly indicated for:
G2M’s Clinical Exome Sequencing (CES) panel is a focused, high-performance complete solution - from DNA to variant report generation tailored for the efficient identification of clinically relevant variants. While conventional CES panels often struggle to balance comprehensive gene content with sequencing performance, the G2M CES panel is meticulously designed to overcome these limitations. The Panel delivers high-quality results that meet the rigorous standards of clinical diagnostic testing, ensuring accuracy you can trust in patient care. With the Clinical Exome Solution by G2M, healthcare providers can more accurately diagnose patients at risk of Mendelian and other genetic disorders, enabling truly personalized and effective treatment plans.
The G2M panel targets approximately 8,124 genes with well-established disease associations, ensuring enriched coverage of medically significant regions with high uniformity and low duplication rates. Curated in accordance with ACMG guidelines, the panel is optimized for diagnostic yield in hereditary disorders and germline cancer risk, offering a cost-effective and clinically actionable alternative to whole exome approaches. With a targeted probe size of 19.6 Mb, the CES panel maximizes sequencing efficiency without compromising sensitivity.

Reference coverage from curated sources (percentage of genes from each database).
The panel adapts a simple, straight-forward workflow enhancing a quick turnaround time. The workflow is divided into:
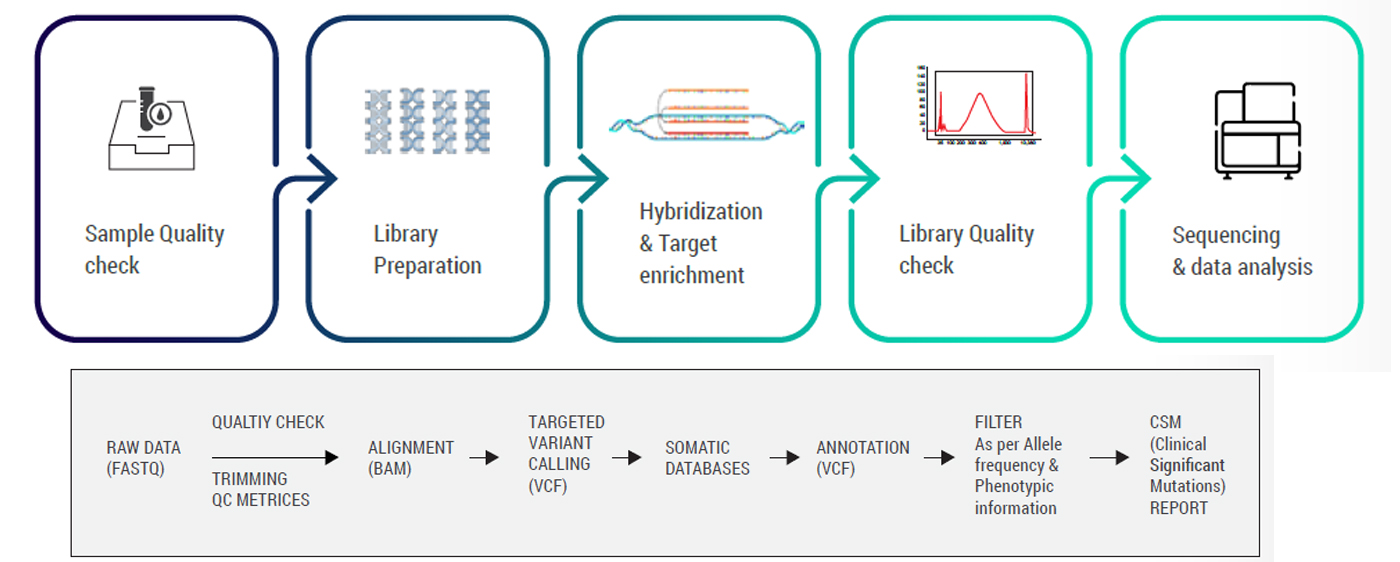
Following raw data generation, variant identification is performed using the GATK best practices framework, beginning with quality assessment via FastQC and alignment to the GRCh38 human reference genome using the BWA aligner. Variants are identified and annotated using advanced tools and databases. Clinically relevant mutations are matched with trusted sources like ClinVar, OMIM, and HPO, while population frequencies and functional impacts are assessed through global genomic databases. The G2M panel targets approximately 8,124 genes with well-established disease associations, ensuring enriched coverage of medically significant regions with high uniformity and low duplication rates. Curated in accordance with ACMG guidelines, the panel is optimized for diagnostic yield in hereditary disorders and germline cancer risk, offering a cost-effective and clinically actionable alternative to whole exome approaches. With a targeted probe size of 19.6 Mb, the CES panel maximizes sequencing efficiency without compromising sensitivity
| Commercial Name | Cat No. | Pack Size | Platform |
|---|---|---|---|
| Clinical Exome Sequencing NGS Test Kit | G710007-1 | 24 T | Illumina |
| G710007-2 | 96 T | Illumina | |
| G710007-3 | 96 T - EZY | Illumina - EZY | |
| Clinical Exome Sequencing NGS Test Kit | G710007-4 | 24 T | MGI |
| G710007-5 | 96 T | MGI | |
| G710007-6 | 96 T - EZY | MGI - EZY | |
| Clinical Exome Sequencing NGS Test Kit | G710007-7 | 24 T | Aviti |
| G710007-8 | 96 T | Aviti | |
| G710007-9 | 96 T - EZY | Aviti - EZY | |
| Clinical Exome Sequencing NGS Test Kit | G710007-10 | 24 T | Thermo |
| G710007-11 | 96 T | Thermo | |
| G710007-12 | 96 T - EZY | Thermo - EZY |
Since its inception in 2016, Genes2me has been constantly striving towards setting a benchmark in the diagnostics space by introducing premium quality (Made in India) diagnostic kits which are CE-IVD, ISO-13485:2016, and ISO 9001:2015 certified, assuring our clients of unparalleled quality and compliance with international standards.
© 2025 Genes2me. All rights reserved.