Around 5-10% of all cancers are hereditary. The most common syndromes are Hereditary Breast and Ovarian Cancer, Lynch syndrome (also called hereditary non-polyposis colorectal cancer), Li-Fraumeni, Cowden syndrome, Familial Adenomatous Polyposis, Von Hippel-Lindau, and Multiple Endocrine Neoplasia (types 1 & 2). Hereditary cancer syndromes are often autosomal dominant, with high penetrance, meaning a single inherited mutation can strongly increase the likelihood of developing cancer; genetic testing and early screening are key to prevention and better outcomes.
Individuals carrying pathogenic variants in cancer-susceptibility genes face a substantially higher likelihood of developing specific cancers compared to those without such mutations, underscoring the importance of early detection and proactive risk management. For instance, a woman carrying a BRCA1 mutation faces up to a 65% lifetime risk of developing breast cancer, compared to just 12% in women without the mutation, highlighting the powerful role genetics play in cancer risk. Similarly, women with an MLH1 gene mutation face a 20% lifetime risk of ovarian cancer, compared to just 3% without the mutation. In the case of an APC gene mutation, the lifetime risk of developing colorectal cancer can reach nearly 100%, while individuals without the mutation have a risk of only about 4%.
Our panel uses advanced Next-Generation Sequencing (NGS) that screens a comprehensive set of genes to identify germline mutations (SNV, InDels & CNV) in DNA associated with inherited cancer syndromes from blood. The panel helps to:
Proven Accuracy of the G2M Hereditary Cancer Panel in Blood Sample Analysis
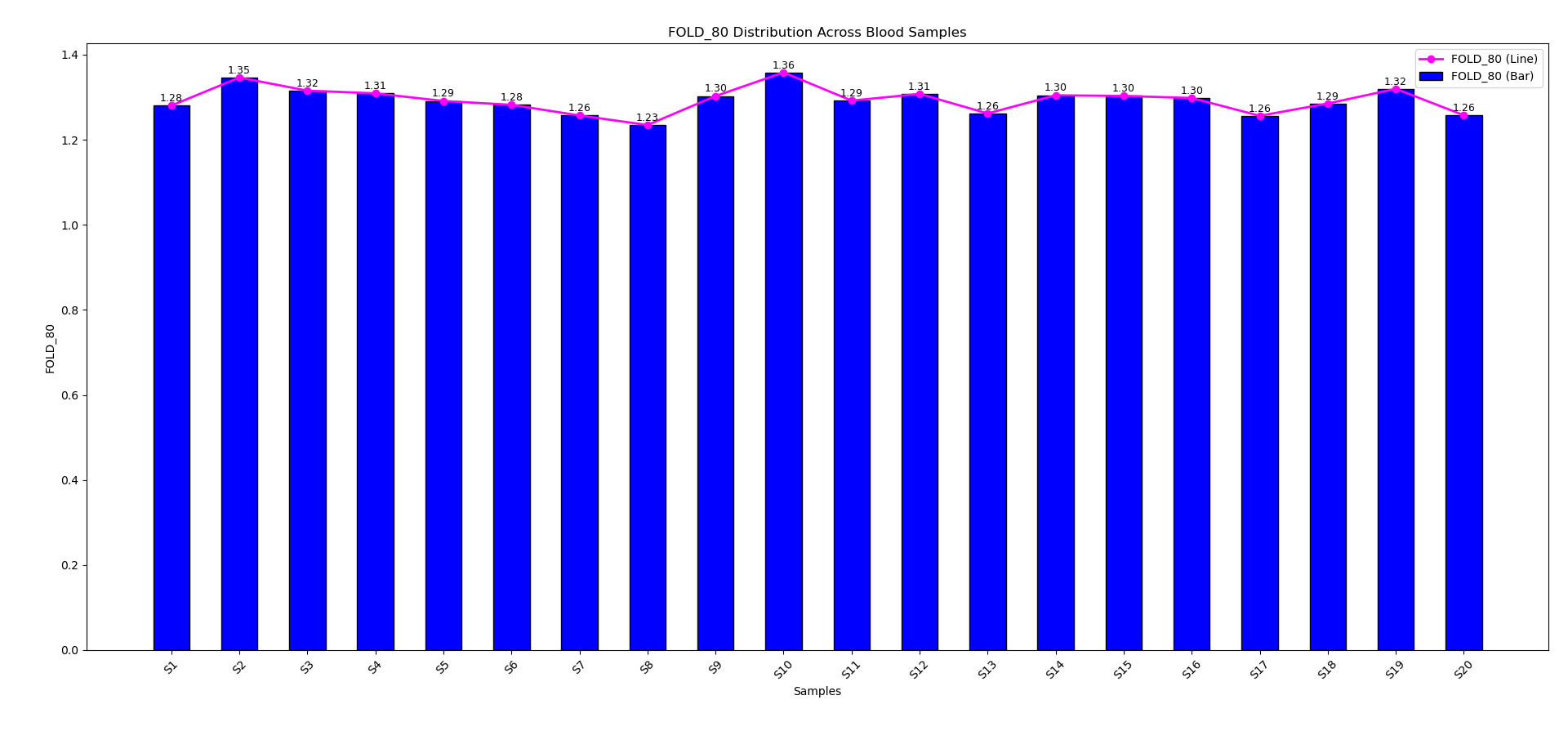
Uniform fold 80 base penalty across blood samples underscores the robust analytical performance of the G2M Common Hereditary Cancer Panel. Fold 80 plots were generated using genomic DNA libraries from blood samples (n=20), enriched with the G2M Common Hereditary Cancer NGS Panel and sequenced on the NovaSeq system using 2 × 150 bp paired-end reads, enabling high-depth, uniform coverage across clinically relevant targets. Fold 80 base penalty across blood-derived samples, below 1.3 demonstrates highly uniform coverage with minimal sequencing bias, reflecting the robustness of the assay design.
On-Target Alignment Across Samples
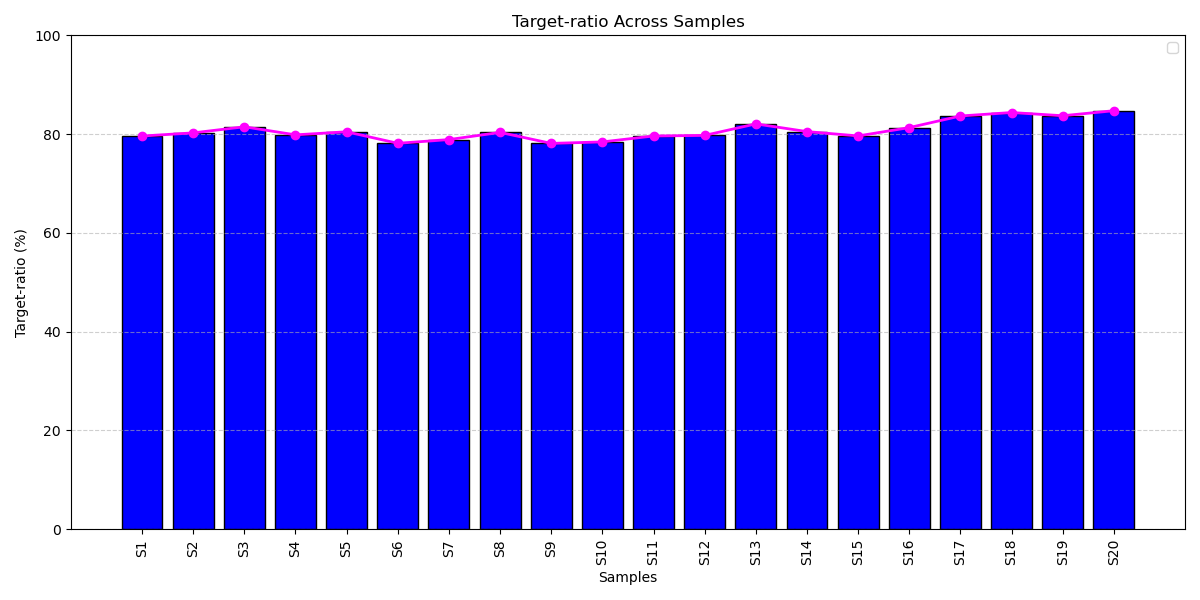
High On-Target Alignment Across Different Cancer Patient Samples: On-target ratios across patient samples consistently exceeded ~80%, highlighting the panel’s optimized design, efficient probe capture, and robust sequencing performance for reliable genomic profiling.
Mean & Median Gene Coverage Supporting Clinical Reliability
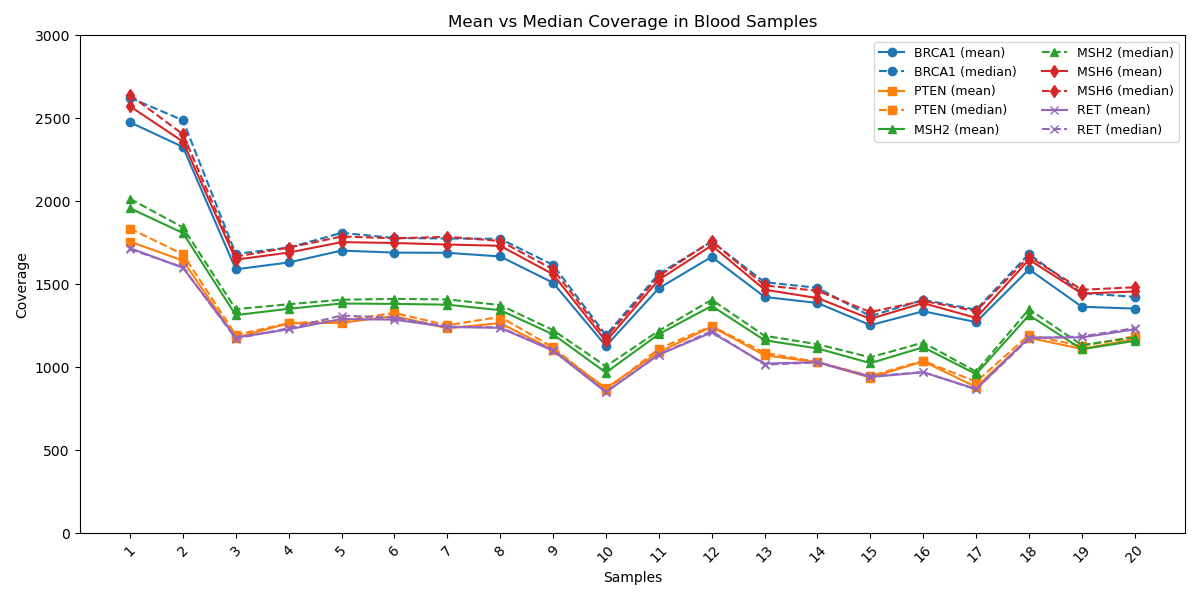
Gene Coverage in Different Cancer Samples: Coverage profiles of key cancer-associated genes (BRCA1, PTEN, MSH2, MSH6, RET) are shown with mean (solid line) and median (dashed line) values. The plot shows a close alignment between mean and median coverage across target genes suggesting a strong indicator of uniform sequencing depth across the specified regions. This highlights the robustness of the workflow, ensuring reliability for routine clinical testing with reliable variant detection across sample types.
Uniform Sequencing Performance for Hereditary Cancer Testing
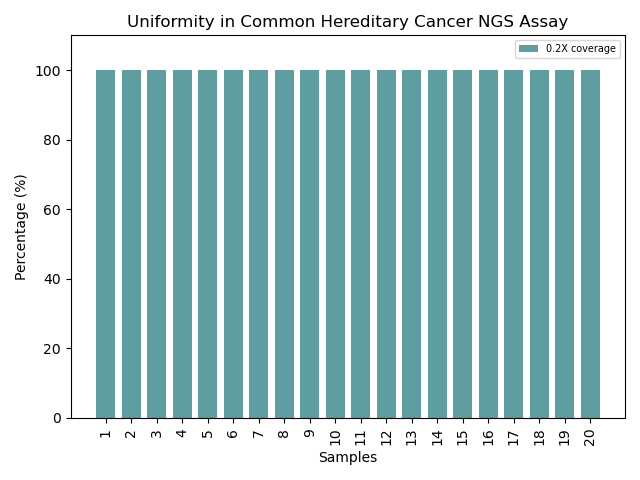
Uniformity That Powers Confidence: Achieving ~100% coverage uniformity at 0.2X across all samples, our optimized probe design ensures consistent enrichment and reliable performance. This translates to high-confidence, reproducible results, empowering every assay with precision and efficiency.
| Commercial Name | Old Cat No. | New Cat No. | Pack Size | Platform |
|---|---|---|---|---|
| Common Hereditary Cancer NGS Test Kit | G2MCHC24001 | G710016-1 | 24 T | Illumina |
| G2MCHC24001 | G710016-2 | 96 T | Illumina | |
| G2MCHC24001 | G710016-3 | 96 T - EZY | Illumina - EZY | |
| Common Hereditary Cancer NGS Test Kit | G2MCHC24001 | G710016-4 | 24 T | MGI |
| G2MCHC24001 | G710016-5 | 96 T | MGI | |
| G2MCHC24001 | G710016-6 | 96 T - EZY | MGI - EZY | |
| Common Hereditary Cancer NGS Test Kit | G2MCHC24001 | G710016-7 | 24 T | Aviti |
| G2MCHC24001 | G710016-8 | 96 T | Aviti | |
| G2MCHC24001 | G710016-9 | 96 T - EZY | Aviti - EZY | |
| Common Hereditary Cancer NGS Test Kit | G2MCHC24001 | G710016-10 | 24 T | Thermo |
| G2MCHC24001 | G710016-11 | 96 T | Thermo | |
| G2MCHC24001 | G710016-12 | 96 T - EZY | Thermo - EZY |
Since its inception in 2016, Genes2me has been constantly striving towards setting a benchmark in the diagnostics space by introducing premium quality (Made in India) diagnostic kits which are CE-IVD, ISO-13485:2016, and ISO 9001:2015 certified, assuring our clients of unparalleled quality and compliance with international standards.
© 2025 Genes2me. All rights reserved.