The Hemat NGS Assay from G2M is designed for comprehensive genomic profiling of hematologic malignancies, enabling high-resolution detection of clinically actionable somatic alterations across leukemias. The assay supports molecular diagnosis, prognostic risk stratification, therapeutic decision-making, and disease classification in accordance with contemporary NCCN, ESMO, and AMP guidelines.
By interrogating DNA mutations, copy number alterations, and both DNA and RNA fusion events, the assay provides a unified solution for routine and advanced clinical genomics applications.
Accurate detection of these variants, including low frequency sub clonal mutations and internal tandem duplications, is critical for risk assessment and therapy selection. The Hemat NGS Assay is engineered to capture difficult genomic regions, including GC-rich, homologous, and repetitive loci that are traditionally challenging for NGS assays.
The Hemat NGS Assay is designed for comprehensive genomic coverage, encompassing 208 genes associated with DNA mutations, 51 DNA fusion genes, and 94 RNA fusion genes within a total target size of approximately 0.7 Mb. The assay covers entire coding sequences along with clinically relevant hotspot regions, enabling robust detection of disease-defining and actionable variants. This integrated assay architecture supports simultaneous interrogation of single nucleotide variants, small insertions and deletions, copy number variations, and both DNA- and RNA-based fusion events within a single, consolidated assay workflow.
The assay employs a hybridization capture–based target enrichment strategy, engineered for efficient and uniform capture of complex genomic regions, including GC-rich, repetitive, and homologous sequences. An optimized hybridization time of approximately 4 hours enables rapid library preparation without compromising to capture specificity or depth of coverage.
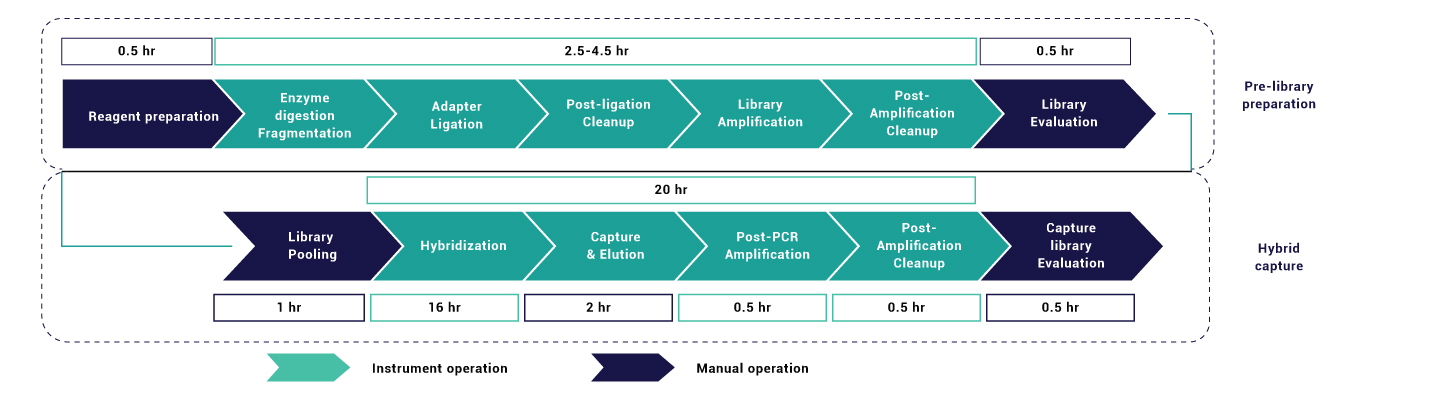
The workflow is designed to be automation-friendly, supporting both manual and automated library preparation formats to accommodate varying laboratory throughput requirements. The assay demonstrates platform-agnostic compatibility and has been validated across commonly used sequencing systems, including Illumina, Element Biosciences, MGI, and Thermo Fisher platforms. This streamlined and flexible workflow ensures high reproducibility, consistent assay performance, and scalability across diverse clinical laboratory settings.
NGS data analysis is supported by GATK-based pipelines integrated with the Cliseq Interpreter Platform, a cloud-based clinical interpretation solution designed to streamline analysis of complex genomic data.
This integrated bioinformatics framework enables standardized, reproducible, and clinically actionable interpretation of leukemia genomic data.
Low frequency variants reveal Leukemia’s true clonal architecture
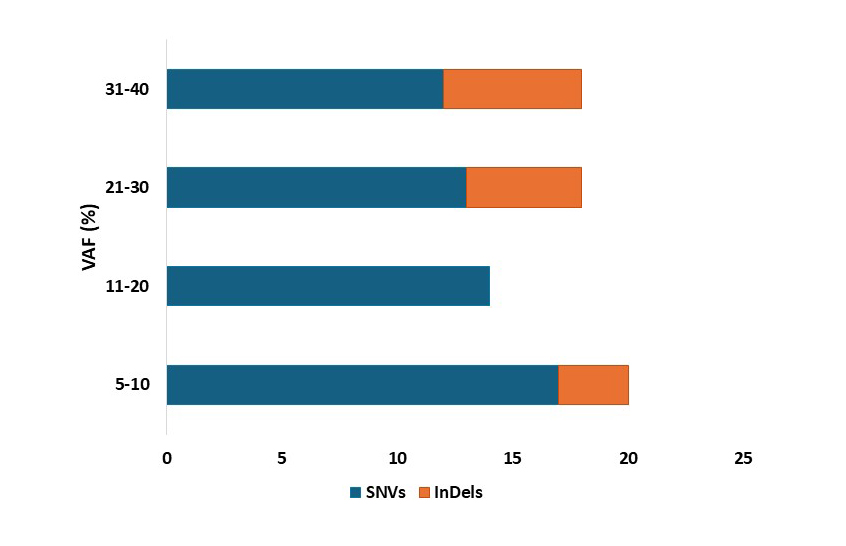
VAF distribution using the G2M Leukaemia kit shows SNVs dominating across all ranges, with notable in Indels presence at low frequencies demonstrating robust detection of low frequency variants for precise driven leukaemia diagnostics.
Mutation profile indicates gene-specific patterns in Leukemia
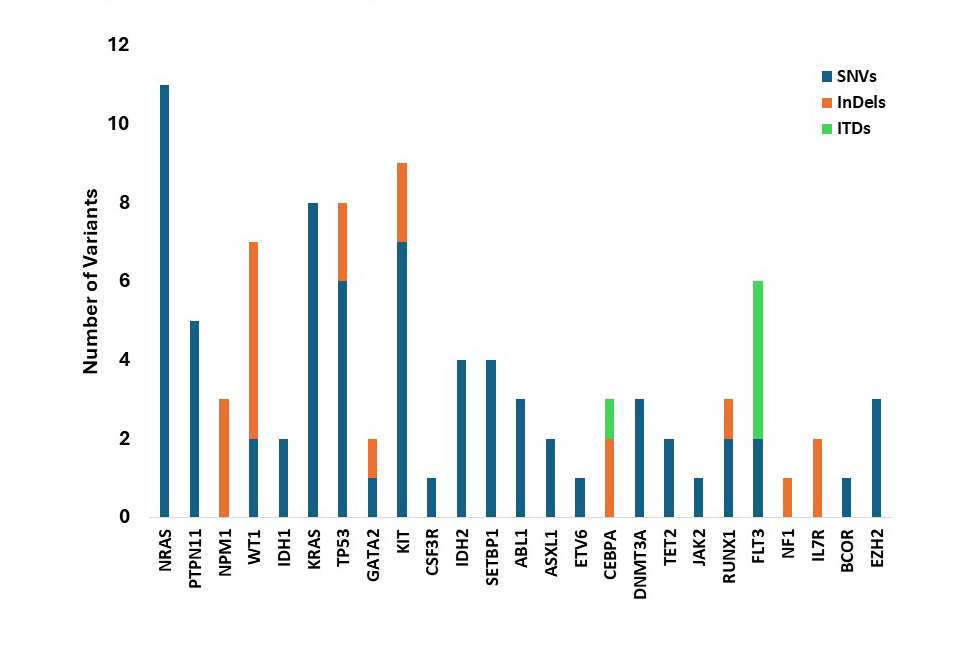
Variant distribution across leukemia patients: SNVs dominate, with NRAS showing the highest count (11). InDels occur in NPM1, WT1, TP53, and other genes, while ITDs are confined to FLT3 and CEBPA.
Precision coverage across all Leukemia samples
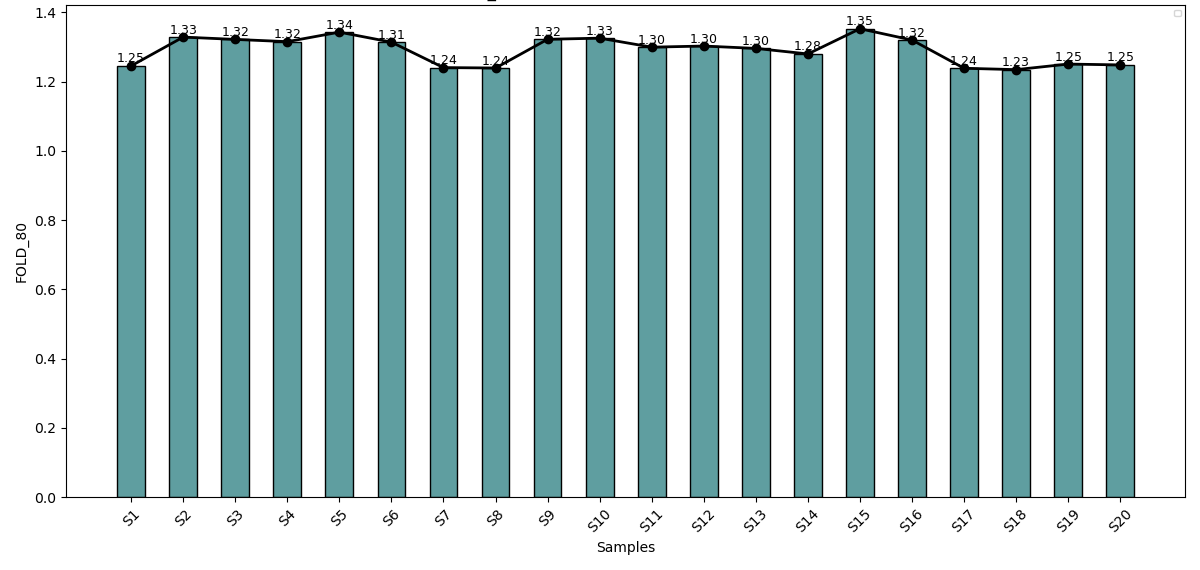
Fold 80 values for blood samples ranged from 1.23–1.35, showing highly uniform coverage with minimal sequencing bias supporting accurate and dependable variant detection.
Fusion driven interaction network exposing critical molecular nodes
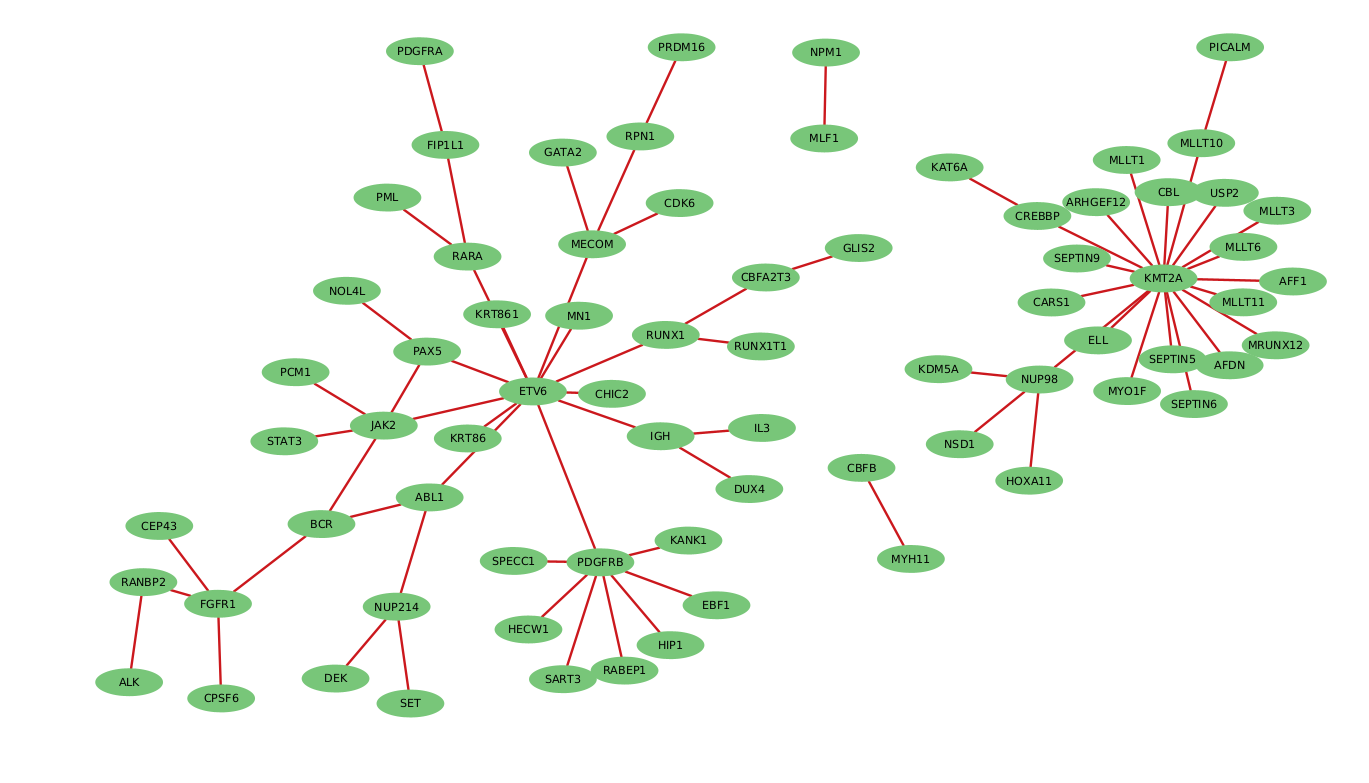
Common fusion partners in G2M Hemat RNA fusion Assay.
RNA fusion detection power in Leukemia
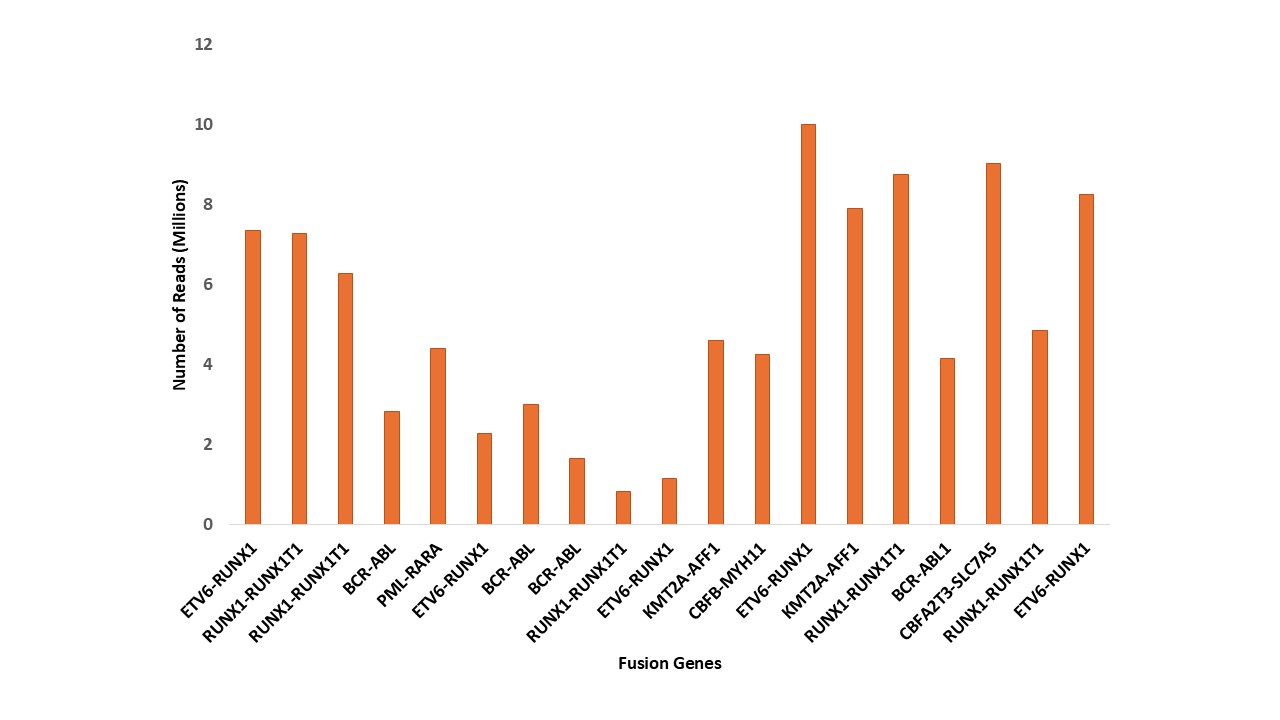
High-resolution profiling of RNA fusion genes in leukemia using our advanced NGS Assay, showcasing detection power across millions of sequencing reads.
VAF distribution across key genes in Leukemia
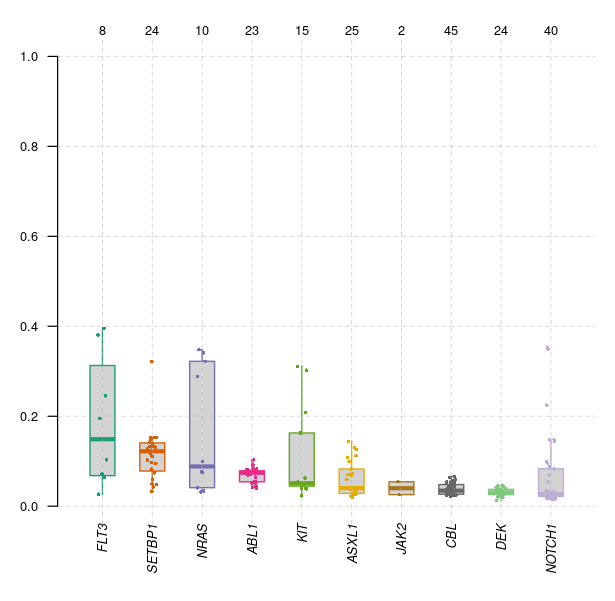
The VAF plot shows the distribution of some of the important genes in 75 myeloid patient samples. Genes like FLT3, SETBP1, and NRAS display higher median VAFs with greater variability, suggesting a higher mutation burden which may reflect a greater impact on disease progression. In contrast, genes like JAK2, CBL, and DEK exhibit lower and more consistent VAFs, indicating a smaller or more stable role in the overall genetic profile.
| Commercial Name | Old Cat No. | New Cat No. | Pack Size | Platform |
|---|---|---|---|---|
| Hemat NGS Test Kit for Leukemia | G2MML28001 | G710017-1 | 24 T | Illumina |
| G2MML28001 | G710017-2 | 96 T | Illumina | |
| G2MML28001 | G710017-3 | 96 T – EZY | Illumina – EZY | |
| Hemat NGS Test Kit for Leukemia | G2MML28001 | G710017-4 | 24 T | MGI |
| G2MML28001 | G710017-5 | 96 T | MGI | |
| G2MML28001 | G710017-6 | 96 T – EZY | MGI – EZY | |
| Hemat NGS Test Kit for Leukemia | G2MML28001 | G710017-7 | 24 T | Aviti |
| G2MML28001 | G710017-8 | 96 T | Aviti | |
| G2MML28001 | G710017-9 | 96 T – EZY | Aviti – EZY | |
| Hemat NGS Test Kit for Leukemia | G2MML28001 | G710017-10 | 24 T | Thermo |
| G2MML28001 | G710017-11 | 96 T | Thermo | |
| G2MML28001 | G710017-12 | 96 T – EZY | Thermo – EZY |
Since its inception in 2016, Genes2me has been constantly striving towards setting a benchmark in the diagnostics space by introducing premium quality (Made in India) diagnostic kits which are CE-IVD, ISO-13485:2016, and ISO 9001:2015 certified, assuring our clients of unparalleled quality and compliance with international standards.
© 2025 Genes2me. All rights reserved.