Assessment of Homologous Recombination Deficiency (HRD) Status and clinical relevance: Homologous Recombination Deficiency (HRD) occurs when the HR repair pathway is compromised due to gene mutations, promoter methylation, or other unknown factors, leading to genomic instability. Tumors with HRD struggle to repair DNA damage effectively, making them vulnerable to targeted therapies. Instead of evaluating individual causes, assessing genomic instability provides a comprehensive measure of HRD status. HRD can be determined by analyzing mutations in HRR pathway genes (such as BRCA1/BRCA2) or by detecting genomic scars and functional assay results.
HRD Status Unlocks Wider Access to PARP Inhibitor Therapy: By blocking PARP-mediated repair, PARP inhibitors trigger synthetic lethality in HR-deficient cancers. HRD testing identifies patients most likely to benefit, going beyond BRCA to include genomic instability markers such as loss of heterozygosity (LOH), Telomeric Allelic Imbalance (TAI) and Large-scale State Transitions (LST).
LOH, detected through copy number analysis of NGS data, indicates the loss or replacement of a parental chromosome segment. In HRD detection, BRCA1/2-specific LOH serves as a critical metric for identifying patients most likely to benefit from PARP inhibitor therapy. Allelic imbalance, reflecting unequal parental DNA contribution after faulty DNA repair, is a key indicator of genomic instability. Telomeric Allelic Imbalance (TAI), extending from the telomere to the sub-telomere, has emerged as a critical metric for assessing HRD status. Genomic instability can also be assessed by measuring Large-Scale State Transitions (LST), which represent chromosomal breaks, creating DNA fragments larger than 10 Mb across the genome.
The HRD Assay from G2M enriches non-exonic, single-nucleotide polymorphism (SNP) based on targeted next generation sequencing. This targets more than 50,000 SNPs enriched across whole genome making it capable of detecting the genomic instabilities and calculate the Genomic Scar Score. These biomarkers LOH (Loss of heterozygosity), TOI (Telomeric imbalances), LSTs (Large scale transitions), can be measured and used to evaluate the HRD Status and Genomic Scar Score (GSS). This helps in maximizing diagnostics insights for clinicians to guide for PARP inhibitors or platinum drugs used in the treatment of various cancers such as ovarian, breast, prostate and pancreatic cancer.
| Number of SNPs: | >51,000 |
| Target Size: | 6.3 Mb |
| Covered Regions: | Whole Coding Sequence, hotspots |
| Biomarkers: | LOH, TAI, LST |
| Sample Type: | FFPE |
| Platform Compatibility: | Illumina, MGI, Thermo Fisher, Element Biosciences |
HRD Score Distribution Highlighting Positive Cutoff for Genomic Instability
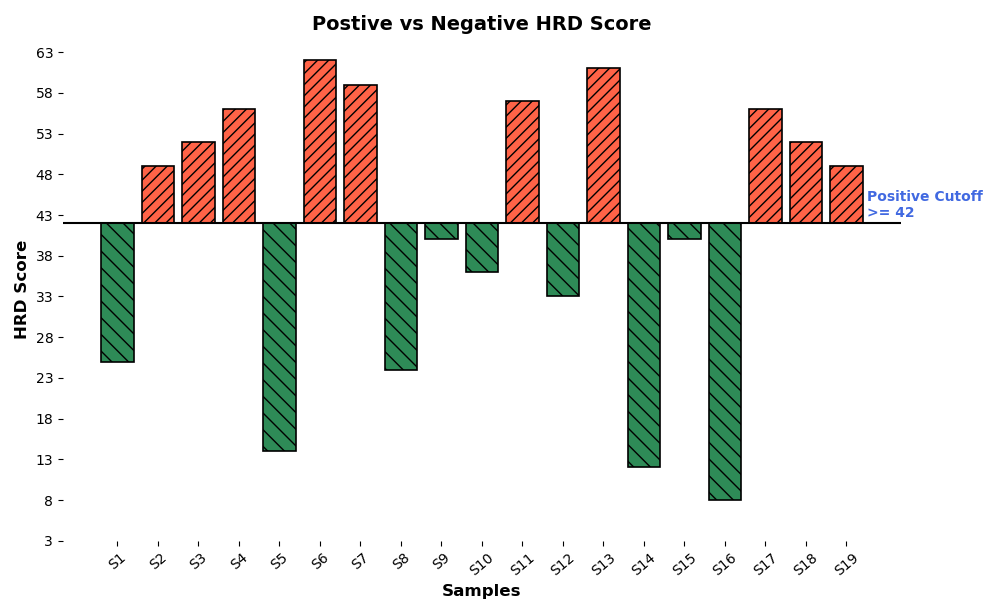
Bar plot illustrating HRD scores for samples on x-axis, categorized as positive ≥42, shown in orange or negative < 42, shown in green. The positive cutoff of 42 highlights samples with significant genomic instability, aiding identification of HRD-positive cases for targeted therapy.
HRD Score Distribution in Matching vs Non-Matching Normal Samples
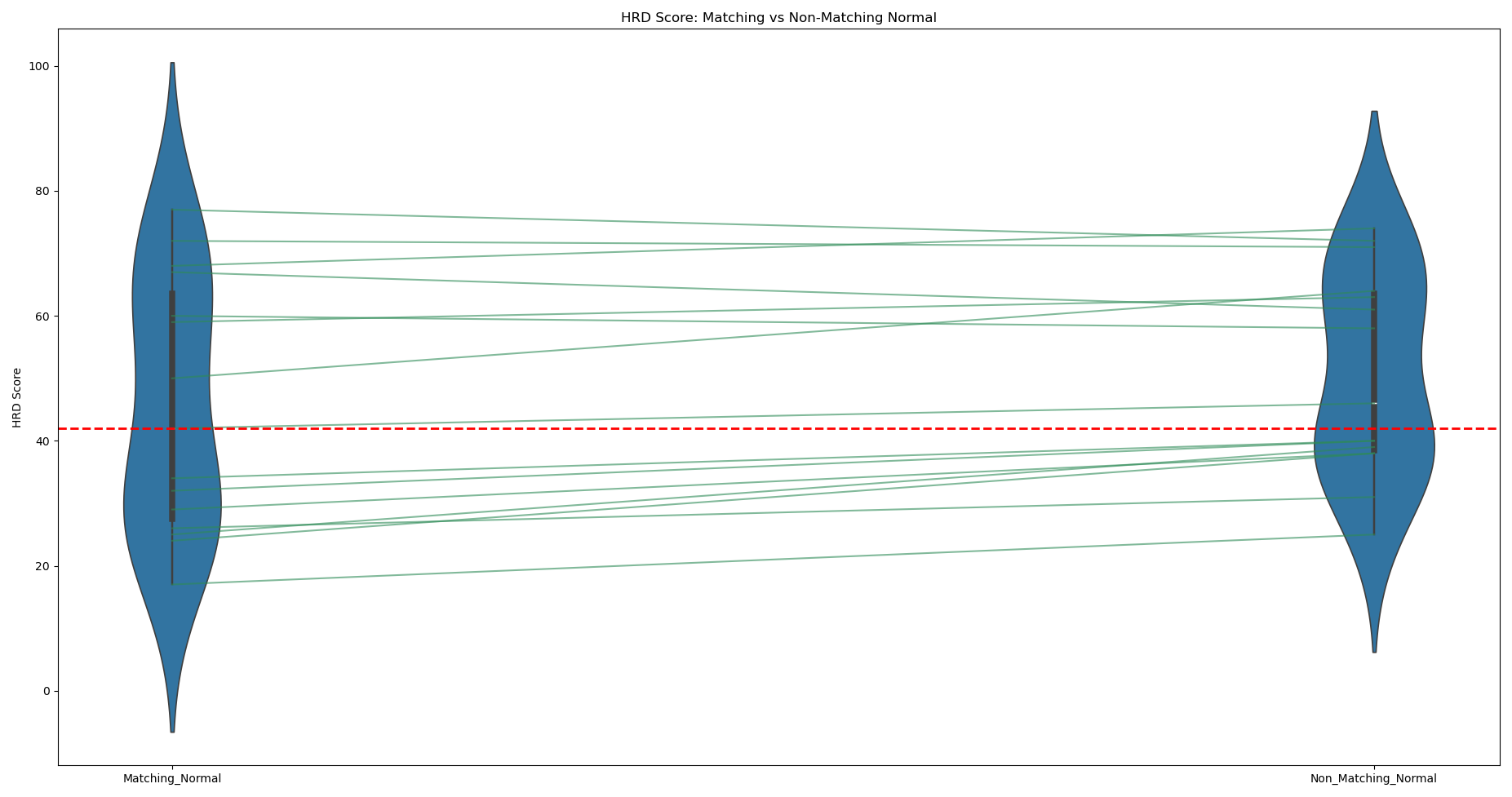
Violin plot illustrating HRD score distribution with the box inside each violin shows the interquartile range (IQR), and the line inside shows the median HRD score. The thin line connects paired observations, and the red dashed line marks the positive cutoff (≥42). This visualization highlights paired differences about how HRD score changes for the same sample when analysed with matching vs non-matching normal.
| Commercial Name | Old Cat No. | New Cat No. | Pack Size | Platform |
|---|---|---|---|---|
| CancerCheck Core NGS Test Kit (HRD) | G2MCC02001 | G710003-1 | 24 T | Illumina |
| G2MCC02001 | G710003-2 | 96 T | Illumina | |
| G2MCC02001 | G710003-3 | 96 T – EZY | Illumina – EZY | |
| CancerCheck Core NGS Test Kit (HRD) | G2MCC02001 | G710003-4 | 24 T | MGI |
| G2MCC02001 | G710003-5 | 96 T | MGI | |
| G2MCC03001 | G710003-6 | 96 T – EZY | MGI – EZY | |
| CancerCheck Core NGS Test Kit (HRD) | G2MCC03001 | G710003-7 | 24 T | Aviti |
| G2MCC03001 | G710003-8 | 96 T | Aviti | |
| G2MCC03001 | G710003-9 | 96 T – EZY | Aviti – EZY | |
| CancerCheck Core NGS Test Kit (HRD) | G2MCC03001 | G710003-10 | 24 T | Thermo |
| G2MCC03001 | G710003-11 | 96 T | Thermo | |
| G2MCC03001 | G710003-12 | 96 T – EZY | Thermo – EZY |
Download useful documents and technical information for the CancerCheck Core HRD Assay
Since its inception in 2016, Genes2me has been constantly striving towards setting a benchmark in the diagnostics space by introducing premium quality (Made in India) diagnostic kits which are CE-IVD, ISO-13485:2016, and ISO 9001:2015 certified, assuring our clients of unparalleled quality and compliance with international standards.
© 2025 Genes2me. All rights reserved.