Liquid Biopsy
ctDNA Breast CancerCancer is one of the leading causes of death worldwide, yet many types still don’t have regular screening programs. Even when screening is available, many people at higher risk find the tests difficult or uncomfortable to follow. Newer blood-based tests, like liquid biopsies, are showing promise for early detection of cancers. Liquid biopsy is a method where circulating tumor DNA (ctDNA) shed from the tumor is extracted and sequenced. This unveils the necessary information regarding cancer causing mutations in the related genes enhancing clinicians to decide the best course of treatment.

G2M offers a panel for liquid biopsy covering major cancer types like Lung, Breast and Colorectal cancer. This panel gives a detailed insight on cancer-related genes to uncover known as well as novel variants for further investigation.
Key features & highlights:
Breast Cancer Detection and Monitoring Powered by Liquid Biopsy and NGS Breast cancer is characterized by the uncontrolled proliferation of malignant epithelial cells within the breast tissue, leading to tumor formation. Although rare in men, it remains predominantly a disease of women and represents a major global health burden. Breast cancer is represented by substantial heterogeneity, encompassing diverse genetic alterations and distinct histopathological subtypes. Despite advances in research, the precise molecular mechanisms driving its initiation and progression remain incompletely understood. The biggest risk factors include simply being female and increasing age. Other factors like family history, gene mutations, race/ethnicity, pregnancy history, breast feeding history and unhealthy lifestyle can also raise the likelihood of developing breast cancer. Research shows that inherited genetic factors contribute to about 5-10% of all breast cancer cases and follow an autosomal dominant inheritance pattern, but in women under 30, they may explain up to 25% of diagnoses. Among these, mutations in the BRCA1 and BRCA2 genes are the most well-recognized causes of hereditary breast cancer risk.
Hereditary breast cancer risk is largely determined by the specific gene affected. Pathogenic variants in BRCA1 and BRCA2 are transmitted in an autosomal dominant manner, whereby a single mutant allele is sufficient to confer increased susceptibility. Although breast cancer predominantly affects women, these germline mutations can be inherited from either the maternal or paternal lineage.
In breast cancer, cell-free circulating tumor DNA (ctDNA) is emerging as a powerful, noninvasive biomarker that may reduce the need for traditional tissue biopsies. Through liquid biopsy, ctDNA shed by tumors into the bloodstream can be analyzed to track genetic changes. Using next-generation sequencing (NGS), even very small amounts of ctDNA can be detected with high accuracy, enabling the identification of both specific gene mutations and broader, genome-wide alterations that drive breast cancer. This helps in guiding targeted therapies and personalized treatment choices. The ctDNA Breast Cancer Panel analyzes key genes linked to tumor growth and treatment response. This table highlights important genes along with the targeted therapies that may be considered.
| Type of Cancer | Gene | Drug |
|---|---|---|
| Breast cancer, Metastatic Castrate Resistant Prostate Cancer, Ovarian Cancer | BRCA1 | Olaparib, Rucaparib, Niraparib + Abiraterone acetate |
| Ovarian Cancer, Breast cancer | BRCA2 | Talazoparib |
| Breast Cancer | ERBB2 | Trastuzumab, Pertuzumab, Ado-trastuzumab emtansine |
| Breast Cancer | ESR1 | Elacestrant (Orserdu) |
Designed for ease and precision, the Genes2me ctDNA Breast Cancer Panel uses a blood/plasma draw to analyze tumor-derived DNA fragments in the bloodstream. This is a hybridization-based solution for targeted sequencing employing NGS. The screening method involves using circulating tumor cells that are used as biomarkers to detect breast cancer. With a fast turnaround time this product provides detection and identifies critical mutations in 63 clinically relevant breast cancer genes spanning 440 Kb of genome size (whole coding sequence) that covers all major mutations like SNV and InDels linked to breast cancer. It is ideal for early diagnosis and detection, prognosis prediction, detecting mutations and structural alterations, minimal residual disease (MRD), tumor mutational burden, and tumor evolution tracking.
| Features | Performance |
|---|---|
| Coverage uniformity (0.2X) | >99% |
| Reproducibility (%) | 96.3 |
| Sensitivity (%) | 96 |
| On Target Ratio (%) | 70-85 |
| Parameter | Description |
| Gene count | 63 (14 DNA fusions) |
| Covered region | Whole CDS |
| Target size | 440 Kb |
| Mutation types | SNV / InDels / CNV |
| Sample type | Blood / Plasma |
| Platform compatibility | Illumina, Thermo Fisher (Ion Torrent), MGI, Element Biosciences |
| Bioinformatics Support: | Primary Analysis: FASTQ to annotated VCF Secondary Analysis: CNVs, SNVs, InDel, Fusions Tertiary Analysis: Clinical interpretation |
High-Fidelity Coverage for Confident Variant Detection
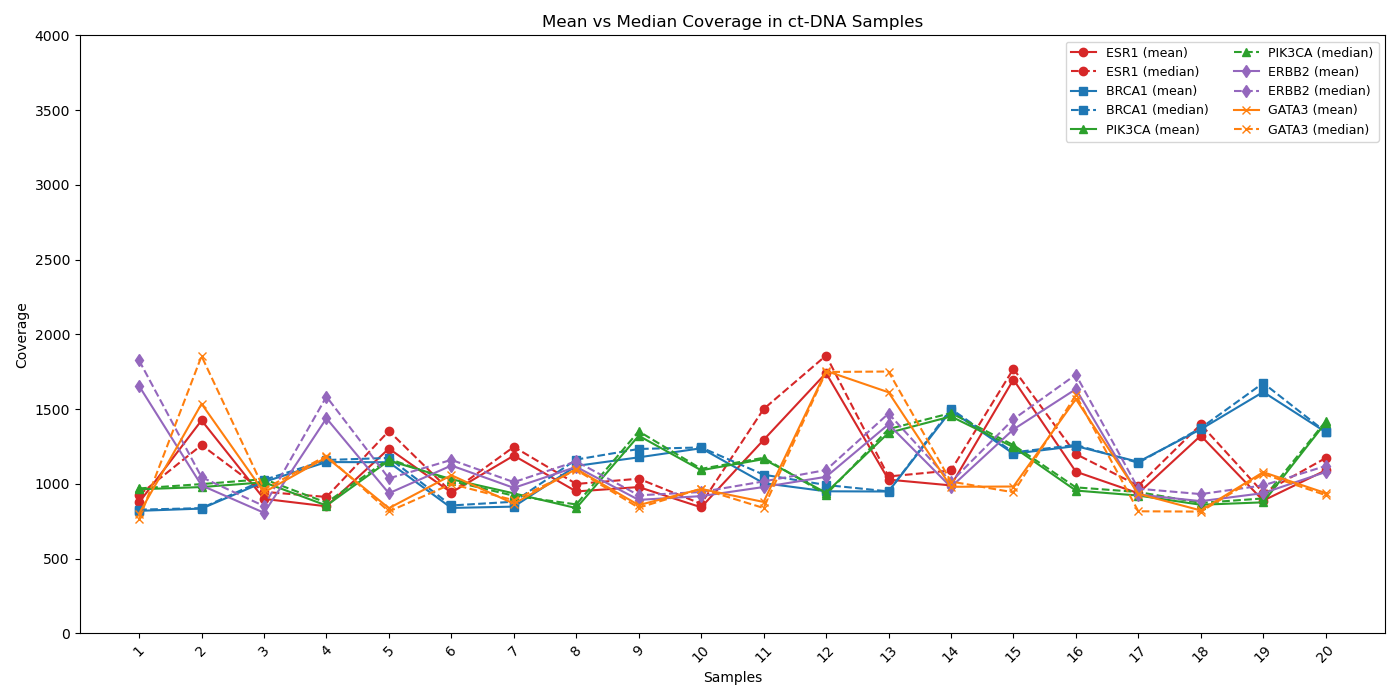
Coverage profiles of critical genes (ESR1, BRCA1, PIK3CA, ERBB2, and GATA3) exhibit strong concordance between mean (solid line) and median (dashed line) depth, demonstrating uniform sequencing and minimal bias across all target regions. This alignment highlights the assay’s robust performance and reliability, ensuring confident results across diverse breast cancer samples.
Mutation Burden Landscape Across Breast Cancer Genes
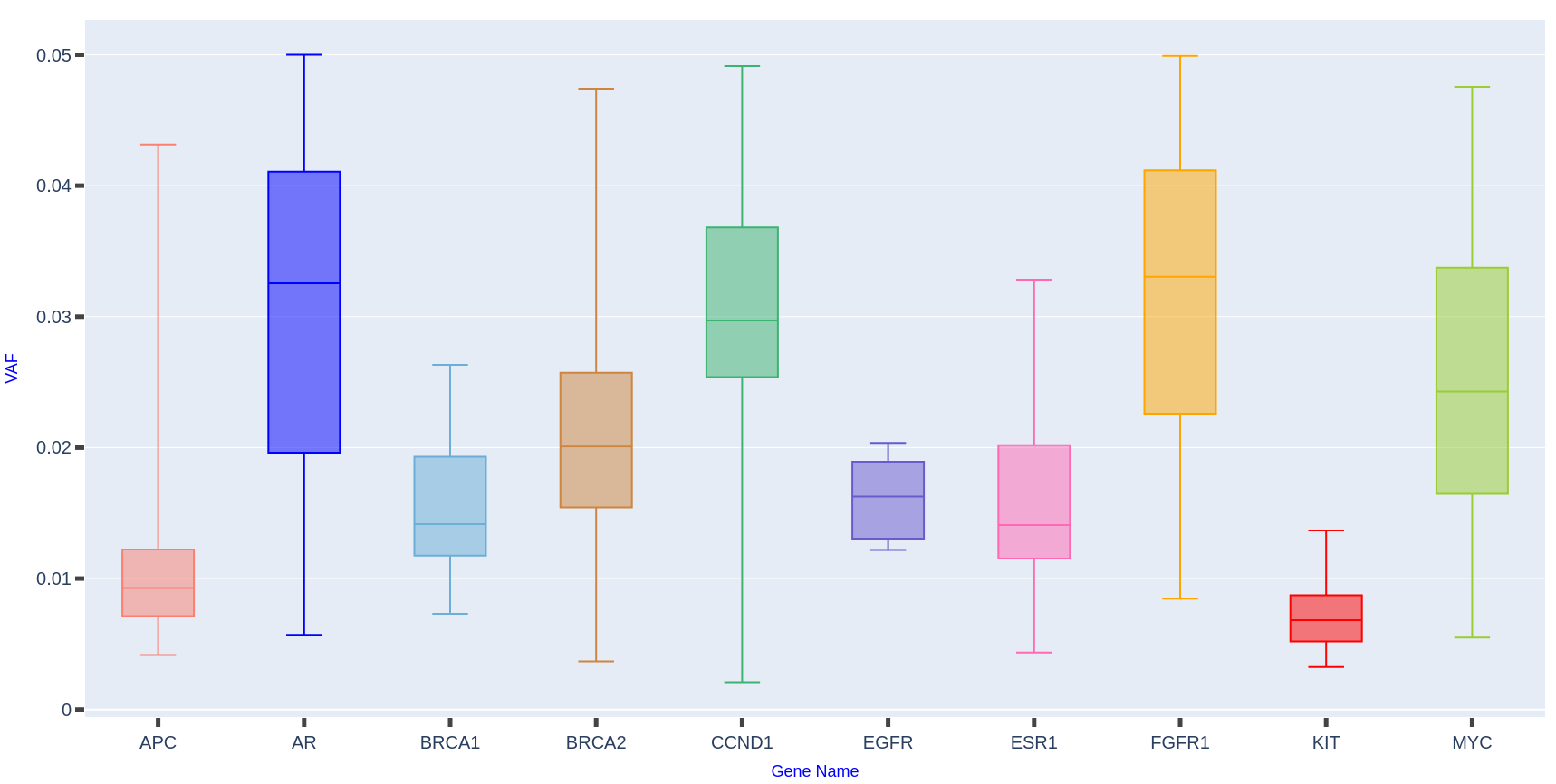
Distribution of variant allele frequencies (VAFs) in 158 breast cancer samples reveals distinct mutation patterns. Genes such as AR, CCND1, and FGFR1 exhibit higher median VAFs with broad variability, indicating a substantial mutation burden and potential influence on disease progression. Conversely, APC, EGFR, and KIT show lower, more uniform VAFs, suggesting a stable or limited role in the overall genomic architecture.
Efficient On-Target Capture for Liquid Biopsy Precision
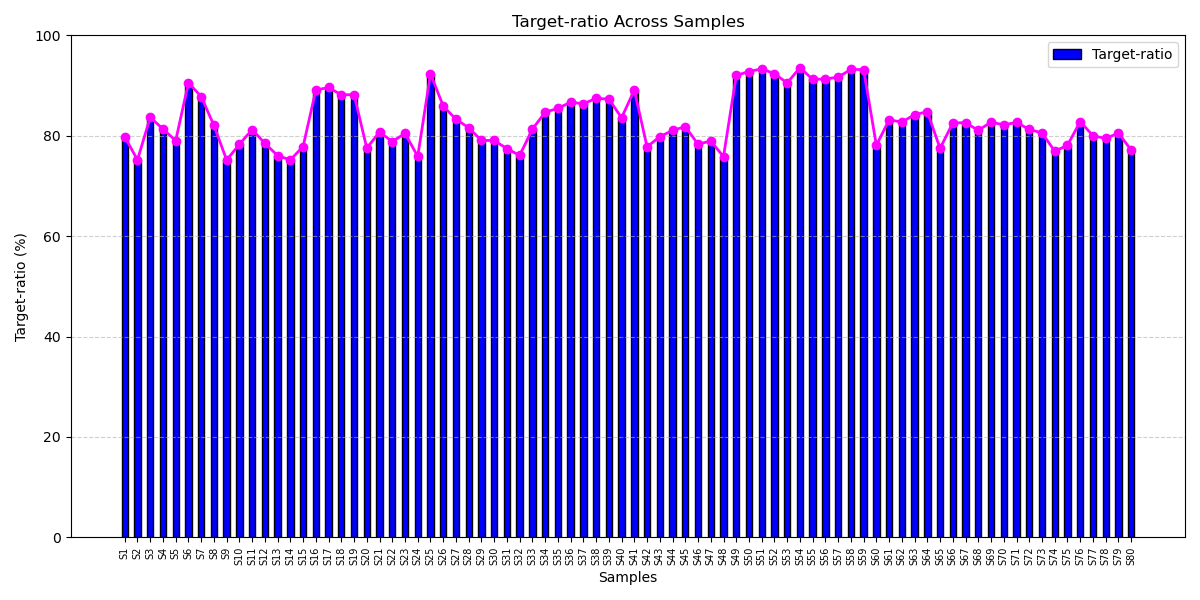
The ctDNA breast cancer panel consistently delivers more than 75% on-target alignment, reflecting its smart design and efficient target capture. This ensures reliable mutation detection from low-input samples, enabling early diagnosis, treatment monitoring, and scalable diagnostic utility.
Uniform BRCA1 Exon Coverage with G2M ctDNA Breast Cancer Panel
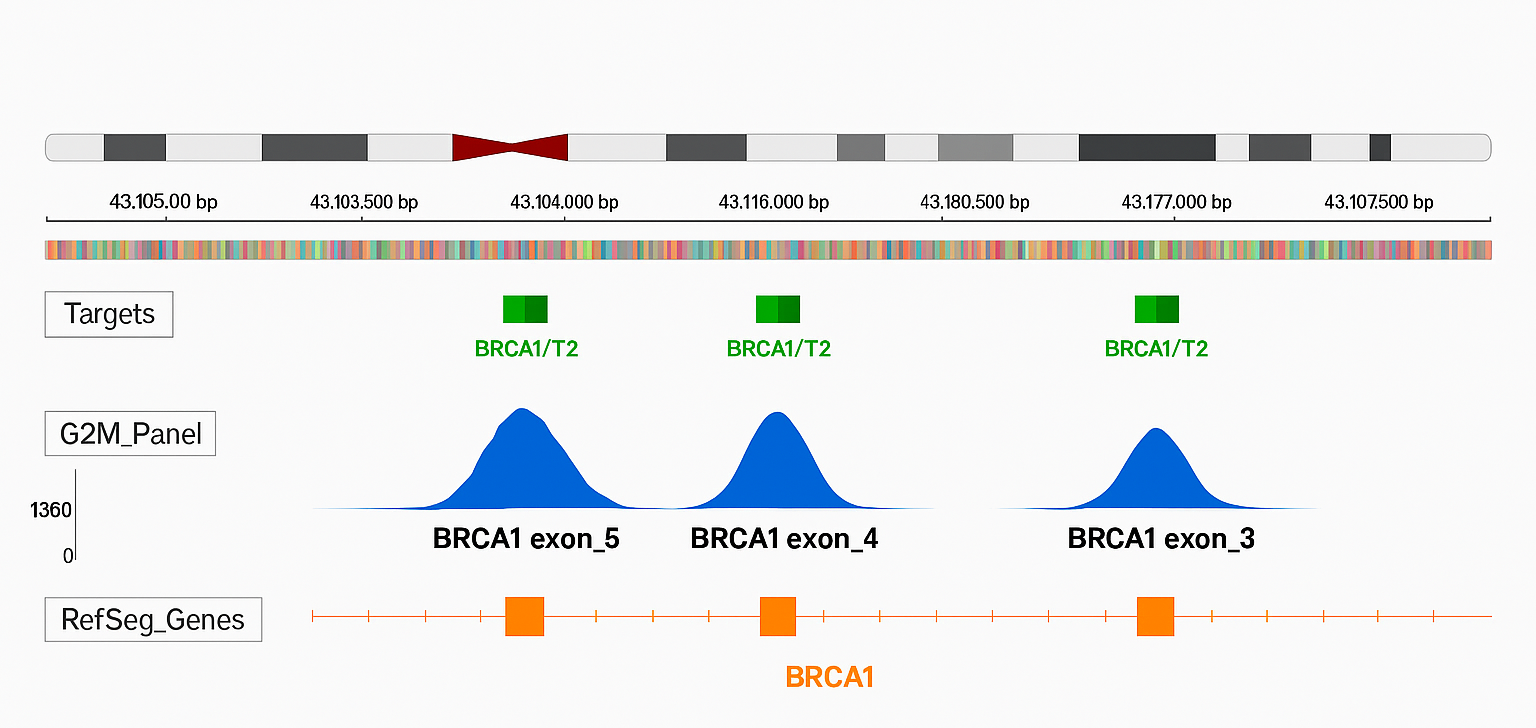
The figure illustrates read coverage for BRCA1 exons 3, 4, and 5. The top panel highlights the BRCA1 target region (green), the middle panel displays per-base coverage depth achieved by the G2M ctDNA Breast Cancer Panel (blue), and the bottom panel represents the gene’s coding regions as annotated by RefSeq (orange). This visualization underscores uniform coverage across clinically significant exons, ensuring reliable variant detection.
Mutation Hotspots Driving Breast Cancer Genomics
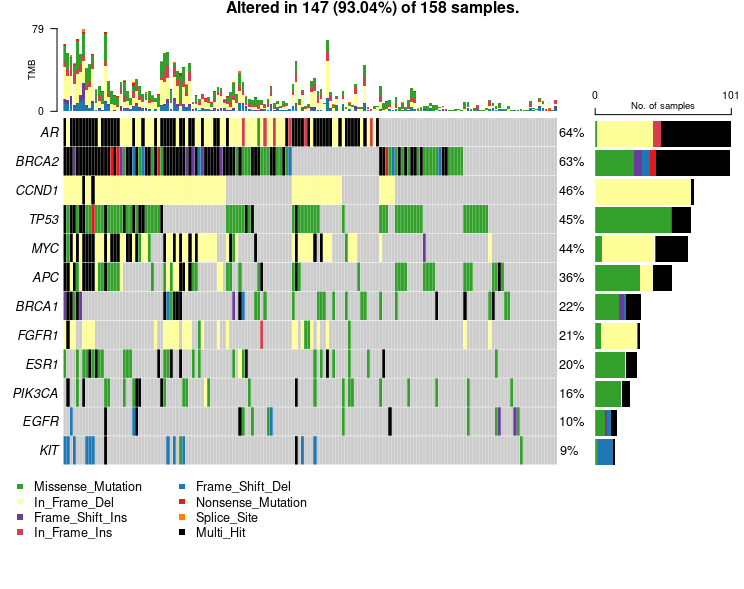
Oncoplot showcasing the 12 most frequently mutated genes in ctDNA breast cancer samples. Missense mutations (green) dominate, followed by in-frame deletions (yellow), while multi-hit events (black) indicate multiple mutation types within the same gene. The right panel summarizes mutation frequency per gene.
| Commercial Name | Old Cat No. | New Cat No. | Pack Size | Platform |
|---|---|---|---|---|
| ctDNA-Breast NGS Test Kit | G2MCTBP12001 | G710011-1 | 24 T | Illumina |
| G2MCTBP12001 | G710011-2 | 96 T | Illumina | |
| G710011-3 | 96 T – EZY | Illumina – EZY | ||
| ctDNA-Breast NGS Test Kit | G2MCTBP12001 | G710011-4 | 24 T | MGI |
| G2MCTBP12001 | G710011-5 | 96 T | MGI | |
| G2MCTBP12001 | G710011-6 | 96 T – EZY | MGI – EZY | |
| ctDNA-Breast NGS Test Kit | G2MCTBP12001 | G710011-7 | 24 T | Aviti |
| G2MCTBP12001 | G710011-8 | 96 T | Aviti | |
| G2MCTBP12001 | G710011-9 | 96 T – EZY | Aviti – EZY | |
| ctDNA-Breast NGS Test Kit | G2MCTBP12001 | G710011-10 | 24 T | Thermo |
| G2MCTBP12001 | G710011-11 | 96 T | Thermo | |
| G2MCTBP12001 | G710011-12 | 96 T – EZY | Thermo – EZY |
Download useful documents and technical information of Liquid Biopsy ctDNA Breast Cancer.
Since its inception in 2016, Genes2me has been constantly striving towards setting a benchmark in the diagnostics space by introducing premium quality (Made in India) diagnostic kits which are CE-IVD, ISO-13485:2016, and ISO 9001:2015 certified, assuring our clients of unparalleled quality and compliance with international standards.
© 2025 Genes2me. All rights reserved.