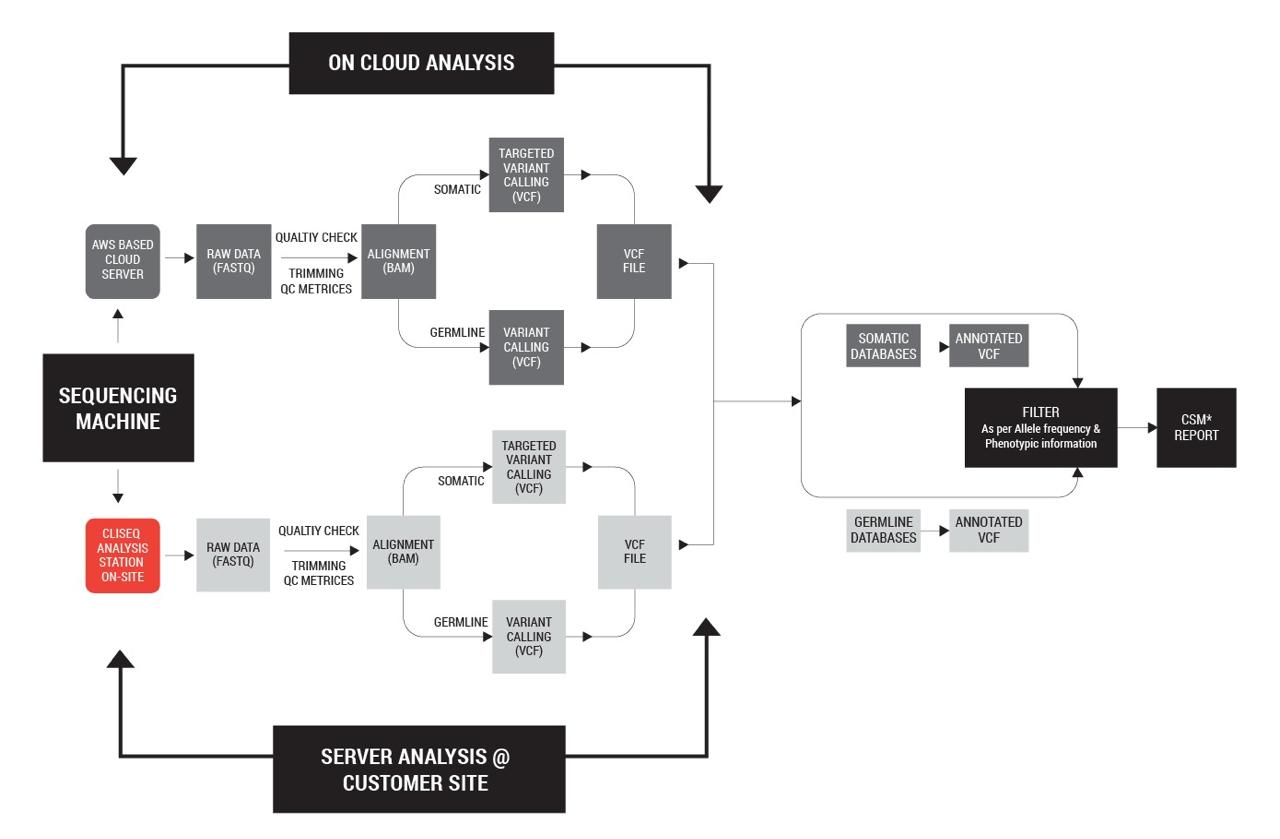
CliSeq Interpreter is a cloud based data analysis software developed by Genes2Me. The software uses automated pipelines for detection of somatic and germline mutations from the raw data generated from major second generation sequencing platforms. FASTQ/ VCF files can be used as input and the pipeline will generate a comprehensive CSM report. CliSeq is GUI driven and user friendly requiring minimum expertise to use. It is Linux based and uses a GPU derived hardware system which makes it faster than many of the CPU only options.